Abstract
Ivermectin Invasive fungal infections cause 1.6 million deaths annually, primarily in immunocompromised individuals. Mortality rates are as high as 90% due to limited treatments. The azole class antifungal, fluconazole, is widely available and has multi-species activity but only inhibits growth instead of killing fungal cells, necessitating long treatments. To improve treatment, we used our novel high-throughput method, the overlap2 method (O2M) to identify drugs that interact with fluconazole, either increasing or decreasing efficacy. We identified 40 molecules that act synergistically (amplify activity) and 19 molecules that act antagonistically (decrease efficacy) when combined with fluconazole. We found that critical frontline beta-lactam antibiotics antagonize fluconazole activity. A promising fluconazole-synergizing anticholinergic drug, dicyclomine, increases fungal cell permeability and inhibits nutrient intake when combined with fluconazole. In vivo, this combination doubled the time-to-endpoint of mice with Cryptococcus neoformans meningitis. Thus, our ability to rapidly identify synergistic and antagonistic drug interactions can potentially alter the patient outcomes.
Invasive fungal infections are an increasing problem worldwide, contributing to 1.6 million deaths annually (Almeida et al., 2019; Bongomin et al., 2017; Brown et al., 2012). These problematic infections are difficult to treat for many reasons. Delayed diagnoses, the paucity and toxicity of antifungal drugs, and the already immunocompromised state of many patients result in mortality rates of up to 90% (Brown et al., 2012; Pianalto and Alspaugh, 2016; Scorzoni et al., 2017). To date, there are only four classes of antifungals, which primarily target the fungal cell envelope (cell wall and plasma membrane) (Coelho and Casadevall, 2016; Odds et al., 2003; Pianalto and Alspaugh, 2016; Scorzoni et al., 2017). The population of immunocompromised individuals is growing due to medical advancements, such as immunosuppression for transplants and chemotherapy. Emerging fungal pathogens are simultaneously increasing in both clinical burden and the number of causal species due to human activity such as agricultural drug use (Berger et al., 2017) and global warming (Almeida et al., 2019; Garcia-Solache and Casadevall, 2010). Thus, the need for more and better antifungal therapeutics is evident. Cryptococcosis is among the most common invasive mycoses, causing 220,000 life-threatening infections and 180,000 deaths annually worldwide (Rajasingham et al., 2017). Cryptococcus neoformans and Cryptococcus gattii are the etiological agents of cryptococcosis, though nearly 95% of cases are caused by C. neoformans (Brown et al., 2012; Maziarz and Perfect, 2016). As C. neoformans is globally distributed throughout the environment, most individuals are exposed by two years of age (Goldman et al., 2001). However, systemic disease primarily occurs in the immunocompromised, particularly those with decreased T helper-1 cell reactions (Maziarz and Perfect, 2016). Accordingly, HIV/AIDS patients account for 80% of cryptococcal cases (Maziarz and Perfect, 2016; Rajasingham et al., 2017). The primary treatment for cryptococcosis involves three different classes of antifungals. Standard care is a combination of amphotericin B (polyene class) and 5-fluorocytosine (5-FC; pyrimidine analog) for two weeks, followed by high dose azole treatment (e.g. fluconazole (FLZ)) for at least 8 weeks, and finally a low dose oral FLZ for at least 6 months (Cox and Perfect, 2018; Mourad and Perfect, 2018). Despite this, mortality rates remain as high as 80% for cryptococcal meningitis (Rajasingham et al., 2017). This is mainly due to the difficulty of obtaining ideal treatment standards. 5-FC is unavailable in 78% of countries, mostly due to licensing issues (Kneale et al., 2016; Mourad and Perfect, 2018). Without the inclusion of 5-FC in the treatment regiment, mortality increases by up to 25% (Kneale et al., 2016). Amphotericin B is administered intravenously, requiring hospitalization. Treatment with amphotericin B is therefore particularly challenging in areas such as sub-Saharan Africa, which has the highest burden of cryptococcal disease (Rajasingham et al., 2017). Due to these therapeutic hurdles, many patients are treated with FLZ alone, which decreases survival rates from 75% to 30% in high burden areas (Kneale et al., 2016). Additional treatment options are thus needed to prevent these unnecessary deaths.
Synergy prediction mutants for fluconazole allow for high-throughput screening of small molecule interactionsWe previously demonstrated that O2M identifies genes whose knockout mutants, termed synergy prediction mutants, exhibit phenotypes that are indicative of synergistic interactions between small molecules (Brown et al., 2014; Wambaugh et al., 2017). O2M identifies synergy prediction mutants by using a chemical-genetics dataset, in which a library of knockout mutants is grown in the presence of >100 small molecules. We calculated quantitative growth scores (slower or faster growth) for each mutant/molecule combination. This large number of phenotypes produces a ‘chemical genetic signature’ for each molecule in the dataset. We then compared the ‘signatures’ for fluconazole and known fluconazole synergizers to identify knockout mutants that exhibit significant phenotypes (i.e. statistically significant slow or fast growth) in all chemical genetic signatures (Brown et al., 2014). The rationale was that similarities between chemical-genetic signatures of known synergistic pairs contain information that is indicative of the synergistic interaction with fluconazole. We term these ‘synergy prediction mutants’ because their knockout phenotype predicts additional fluconazole-synergizing molecules. We then identified genes whose mutant exhibited significant growth scores (|Z| > 2.5) when compared to wild-type growth for fluconazole and each of its known synergizers. We compared the significant gene set for fluconazole and each synergizer and identified common genes. We called these ‘synergy prediction mutants’ (Figure 1A). Since fluconazole and its known synergizers induce a growth phenotype from these mutants, we hypothesized that any molecule eliciting the same growth phenotype would also act synergistically with fluconazole. This was completed and tested in our previous publication (Brown et al., 2014) using FLZ and its known synergistic interacting partners fenpropimorph and sertraline (Jansen et al., 2009; Zhai et al., 2012). Our work identified three synergy prediction mutants (cnag_00573Δ, cnag_03664Δ, and cnag_03917Δ) (Brown et al., 2014). Since each chemical-genetic signature contains phenotypes from ~1400 gene knockouts, there does not have to be considerable overlap between chemical-genetic signatures to identify synergy prediction mutants.
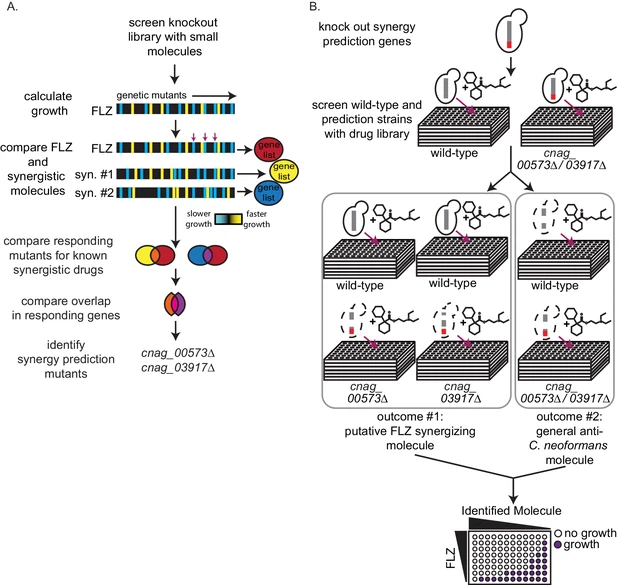
Here we use synergy prediction mutants to rapidly screen for small molecules that synergize with fluconazole and can be quickly moved into clinical use. CNAG 00573 encodes a NADH dehydrogenase (Janbon et al., 2014), CNAG 03664 encodes NIC1, a high-affinity nickel-transporter (Singh et al., 2013), and CNAG 03917 encodes a nuclear pore complex protein homologous to Nup75 (Liu et al., 2008; Stajich et al., 2012). Using these gene mutants, we performed a high-throughput screen for synergistic interactions. Our assay is simple: differential growth between wild-type and synergy prediction mutants is indicative of a synergistic interaction with FLZ or any other starting drug. It does not require multi-drug assays, as the ‘synergy prediction mutant’ substitutes for one of the small molecules in the interaction, phenocopying the FLZ-small molecule interaction to produce synthetic lethality. We screened the Microsource Spectrum Collection, a small-molecule library of 2000 compounds enriched for FDA-approved molecules. We grew C. neoformans wild-type and synergy prediction mutants (cnag_00573Δ and cnag_03917Δ) in the presence of each small molecule (1 µM), identifying those that caused a significant difference in growth between the wild-type and both synergy prediction mutants after 48 hr of growth (Figure 1B). The mutant cnag_03664Δ was not used due to its inherent slow growth. The small molecule concentration of 1 µM gave the lowest false discovery rate when testing molecules known to synergize or not synergize with fluconazole. Using these synergy prediction mutants, we identified 313 putative FLZ synergistic molecules (Supplementary file 1).
We validated potential synergistic interactions in checkerboard assays, for which serial dilutions of each drug are crossed in a 96-well plate (Figure 1B). Synergistic interactions are defined as a ≥ 4 fold decrease in the minimum inhibitory concentration (MIC) of each small molecule in the pair, resulting in a fractional inhibitory concentration index (FICI) of ≤0.5 (Johnson et al., 2004; Odds, 2003). We tested the 129 molecules with single agent efficacy against C. neoformans growth in the preferred checkerboard assay. We found that 40 molecules were synergistic with FLZ, meaning 31% of these molecules were correctly predicted by O2M (Figure 2A). However, checkerboard assays require that both small molecules in the pair are able to inhibit growth of C. neoformans individually, which was not the case with all our putative synergistic molecules. In those cases, we performed Bliss Independence, which identifies whether molecules enhance the action of FLZ (Tang et al., 2015). In a 96-well plate, we created a gradient of FLZ combined with 10 µM and 100 nM concentrations of the 55 small molecules that could not inhibit C. neoformans alone. We found 6 of these molecules enhanced the action of FLZ at both 10 µM and 100 nM concentrations and were deemed synergistic (Figure 2—figure supplement 1). The FLZ-synergistic molecules belonged to a wide range of bioactive categories including antidepressants, adrenergic agonists, as well as antiinfectives (Figure 2B and Table 1).

credited to e life
Comments